Temporally averaged Level 3 data from TROPOMI L2 NO2 observations¶
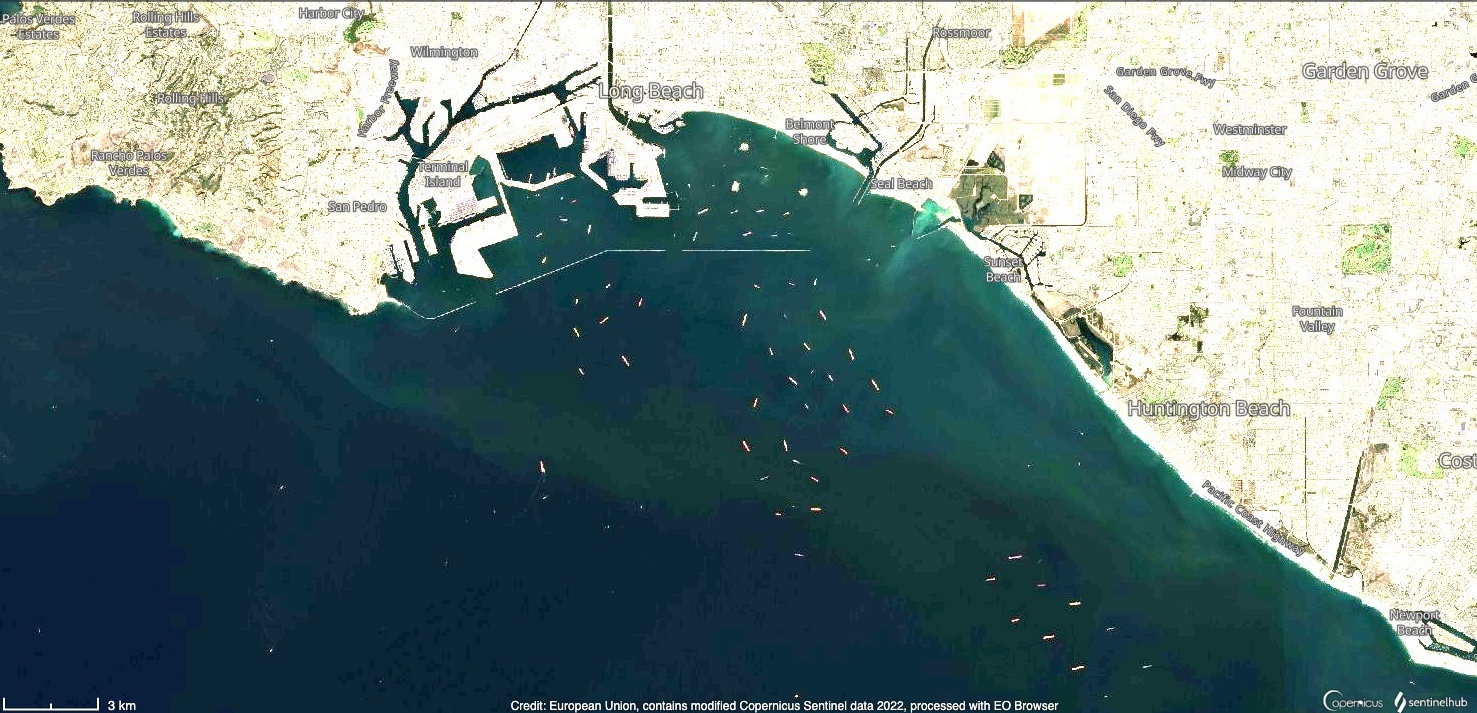
This use case demonstrates how to create a gridded and temporally averaged level 3 data from several level 2 TROPOMI/S5P NO2 files. In this example the focus is on the Los Angeles Port area. In October 2021 major ports in the US faced record backlogs due to rapidly increased amount of cargo. Large container ships were staying for days outside the ports waiting for their turn to unload. The above Sentinel 2 image shows number of container ships in the Los Angeles coast on 15th October. Ships emit NO2, and it can be expected that in this kind of backlog situation these large container ships could potentially increase the NO2 concentrations in the area. According to NASA's science news the unusual shipping activity might indeed play a role in elevated NO2 concentrations, that have been observed in major port areas. In this use case it is demonstrated how TROPOMI NO2 observations can be averaged for one-week period (11-17th October 2021) using harp. It is also demonstrated how the averaged data can be filtered according to surface wind speed.
1. TROPOMI Level 2 NO2 product ¶
TROPOMI provides observations of total and Tropospheric column concentrations of NO2. In this use case also information on wind is used. The northward and eastward wind components from ECMWF model at 10 meter height have been provided in TROPOMI NO2 files since August 2019. For more information on the TROPOMI NO2 product can be found from the following documents:
2. Python packages for the notebook ¶
This example uses the same Python packages as Use Case 2. In this example the basemap for plotting is retrieved from cartopy Stamen map tiles, therefore import cartopy.io.img_tiles as cimgt is added when importing packages.
import harp
import numpy as np
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.io.img_tiles as cimgt
from cmcrameri import cm
import eofetch
3. Downloading TROPOMI NO2 files (optional) ¶
The TROPOMI NO2 level 2 data used in this notebook is obtained from the Sentinel-5P Pre-Operations Data Hub. For Sentinel-5P each level 2 file contains information from one orbit. This notebook uses TROPOMI NO2 data between 11th and 17th October 2021. The orbits that cover the area of interest, i.e. port of Los Angeles, and used in this example are:
- 20706, 20720, 20734, 20748, 20749, 20762, 20763, 20777, 20791
filenames = [
"S5P_RPRO_L2__NO2____20211011T200531_20211011T214700_20706_03_020400_20221105T135504.nc",
"S5P_RPRO_L2__NO2____20211012T194622_20211012T212752_20720_03_020400_20221105T141035.nc",
"S5P_RPRO_L2__NO2____20211013T192714_20211013T210844_20734_03_020400_20221107T063813.nc",
"S5P_RPRO_L2__NO2____20211014T190806_20211014T204936_20748_03_020400_20221105T142626.nc",
"S5P_RPRO_L2__NO2____20211014T204936_20211014T223105_20749_03_020400_20221105T142627.nc",
"S5P_RPRO_L2__NO2____20211015T184858_20211015T203028_20762_03_020400_20221105T143409.nc",
"S5P_RPRO_L2__NO2____20211015T203028_20211015T221157_20763_03_020400_20221105T143410.nc",
"S5P_RPRO_L2__NO2____20211016T201119_20211016T215249_20777_03_020400_20221105T143432.nc",
"S5P_RPRO_L2__NO2____20211017T195211_20211017T213341_20791_03_020400_20221105T144233.nc",
]
eofetch.download(filenames)
Filtering and gridding of TROPOMI Level 2 data is done by defining specific operations for the harp.import_product() function. The operations for this is use case will follow closely the examples in Use Case 1 and Use Case 2. The main differences are NO2-specific quality filtering and deriving surface wind speed and filtering data according to that:
1) tropospheric_NO2_column_number_density_validity>75 to consider only pixels that are of high quality. The quality criteria for NO2 product can be found from Product Readme File for NO2
2) derive(surface_wind_speed {time} [m/s]): derive wind speed from meridional and zonal surface wind velocity components.
3) surface_wind_speed<5: consider only data where wind speed is below 5 m/s.
4) keep(latitude_bounds,longitude_bounds,datetime_start,tropospheric_NO2_column_number_density, surface_wind_speed): keep only selected variables from the original netcdf files. Note that in this example we use the tropospheric NO2 column data.
5) bin_spatial(nlat,latoff,latincr,nlon,lonoff,lonincr): define the common Level 3 grid. Detailed description for bin_spatial() input is found from Use_Case_1, Step4. For this use case we create a 0.02 deg lat, lon grid, where the latitude and longitude offsets are 33.5N, -118.5W, respectively, and the gird extends about 1 deg to the east and north from the offset. Thus bin_spatial() input parameters are:
bin_spatial(51,33.50,0.02,51,-118.5,0.01)
6) derive(tropospheric_NO2_column_number_density [Pmolec/cm2]): convert the NO2 units to 10^15 molec/cm^2.
operations = ";".join([
"tropospheric_NO2_column_number_density_validity>75",
"derive(surface_wind_speed {time} [m/s])",
"surface_wind_speed<5",
"keep(latitude_bounds,longitude_bounds,datetime_start,datetime_length,tropospheric_NO2_column_number_density, surface_wind_speed)",
"derive(datetime_start {time} [days since 2000-01-01])",
"derive(datetime_stop {time}[days since 2000-01-01])",
"exclude(datetime_length)",
"bin_spatial(51,33.50,0.02,51,-118.5,0.02)",
"derive(tropospheric_NO2_column_number_density [Pmolec/cm2])",
"derive(latitude {latitude})",
"derive(longitude {longitude})",
"count>0"
])
B. Reduce operations for temporal averaging ¶
Reduce operations is defined in a similar manner than the operations string, the explanations can be found from e.g. Use Case 2.
reduce_operations=";".join([
"squash(time, (latitude, longitude, latitude_bounds, longitude_bounds))",
"bin()"
])
C. Applying HARP import to multiple NO2 files ¶
To apply the harp import function to all the needed files, wildcard * can be used e.g. as follows:
files_in="S5P_RPRO_L2__NO2____202110*.nc"
and finally run harp_import():
mean_no2=harp.import_product(files_in, operations, reduce_operations=reduce_operations)
D. Saving data as netcdf ¶
You can save the merged level 3 data into a new netcdf file using harp.export_product:
harp.export_product(mean_no2, 's5p-NO2_L3_averaged_11-17_Oct2021.nc')
5. Plotting NO2 data on a map ¶
Now that the averaged level 3 NO2 product is given in 2D, we can use cartopy and pcolormesh function to visualise the merged NO2 data on a map. This use case follows closely the example in Use Case 1 to define the NO2 data field, latitude, longitude, and colormap:
no2 = mean_no2.tropospheric_NO2_column_number_density.data
no2_description = mean_no2.tropospheric_NO2_column_number_density.description
no2_units = mean_no2.tropospheric_NO2_column_number_density.unit
gridlat = np.append(mean_no2.latitude_bounds.data[:,0], mean_no2.latitude_bounds.data[-1,1])
gridlon = np.append(mean_no2.longitude_bounds.data[:,0], mean_no2.longitude_bounds.data[-1,1])
colortable = cm.roma_r
vmin = 0
vmax = 14
Next the base map is defined, and NO2 data is plotted. In this example a map tile from Stamen is retrieved as a basemap. The style is set as "toner-lite". The map lat and lon extent are set with boundaries-variable, and the map zoom level is set to 10. The alpha argument in plt.pcolormeshdefines the transparency of the NO2 data on top of the basemap.
# Setting basemap
boundaries=[-118.5, -117.5, 33.5, 34.0]
fig = plt.figure(figsize=(10,10))
bmap=cimgt.OSM()
ax = plt.axes(projection=bmap.crs)
ax.set_extent(boundaries,crs = ccrs.PlateCarree())
zoom = 10
ax.add_image(bmap, zoom)
# Adding NO2 data
img = plt.pcolormesh(gridlon, gridlat, no2[0,:,:], vmin=vmin, vmax=vmax, cmap=colortable, transform=ccrs.PlateCarree(),alpha = 0.55)
ax.coastlines()
cbar = fig.colorbar(img, ax=ax,orientation='horizontal', fraction=0.04, pad=0.1)
cbar.set_label(f'{no2_description} [{no2_units}]')
cbar.ax.tick_params(labelsize=14)
plt.show()
As the figure shows, NO2 concentrations are much higher at the Los Angeles downtown area than over the ocean, as expected, and it is difficult to say whether the ships have affected to the NO2 concentrations over the sea. There are many factors, e.g. meteorology, that affect the calculated weekly NO2 average, and therefore comparisons with corresponding time period from previous years is not straightforward. Filtering the data with respect to the surface wind speed (include only "weak" wind cases), somewhat reduces the meteorological effects. A reference data could be created averaging the same time period e.g. from 2019 and 2018. It should be noted, however, that the surface wind components are included in the TROPOMI NO2 data only from August 2019, and thus filtering according to wind speed is possibly with data only after that.