Demonstration of QDOAS and BeAMF¶
This use case demonstrates the use of QDOAS and BeAMF functionalities that are included in the newest version of Atmospheric Virtual Laboratory. QDOAS performs DOAS retrievals of trace gases from spectral measurements whereas BeAMFis a tool to calculate the Air Mass Factor (AMF) to make the conversion from trace gas slant column density (SCD) to vertical column density (VCD).
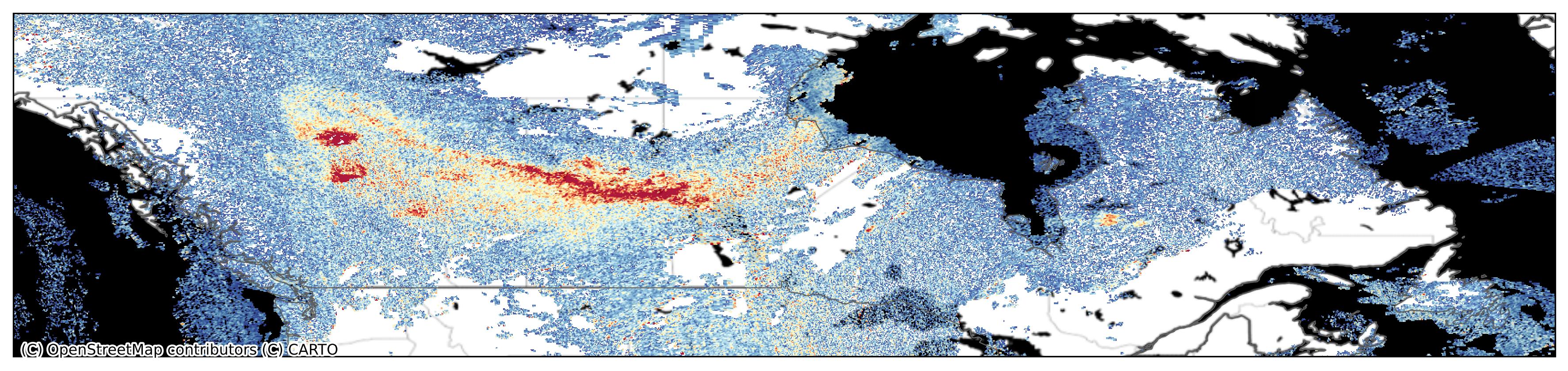
In this example formaldehyde (HCHO) vertical column density from TROPOMI (S5P) L1B data is retrieved using QDOAS and BeAMF. The interest is to observe enhanced levels of HCHO in Canada in June 2023, when large parts of the country were affected by a series of wildfires.
Table of contents¶
1. Initial preparations ¶
A. Input files
In this tutorial the input data is based on the TROPOMI L1B radiance spectra measured on 8 June 2023 (orbit 29289). In addition, the corresponding operational S5P HCHO L2 file is needed to obtain auxiliary input parameters for BeAMF calculations.
This notebook and BeAMF requires the following files:
TROPOMI L1B data: S5P_OFFL_L1B_RA_BD3_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc, S5P_OFFL_L1B_IR_UVN_20230608T005615_20230608T023745_29278_03_020100_20230608T042535.nc
Configuration file and additional data for QDOAS: S5P_TROPOMI_HCHO_ISFRfit_qdoasconfig_radasref.xml and the corresponding data folder that contains input needed for the DOAS fit.
Configuration file for the BeAMF tool: harp_hcho.json
Box-AMF and Radiance Look-up-table (LUT): LUT_AMF_I_300nm_500nm_S1_US_small_new.nc
Operational TROPOMI L2 HCHO file: S5P_OFFL_L2__HCHO___20230608T193244_20230608T211414_29289_03_020401_20230610T184103.nc
The QDOAS configuration file uses the so-called radiance-as-reference setting: because formaldehyde is a weak absorber, relatively large offsets in the retrieved slant columns are generally observed. These can be mitigated by an active offset correction or by normalizing the spectral data by an average background radiance field, included here in the file data/rar_20230608.nc. Retrieving formaldehyde from measurements from a few days earlier or later can safely use the same background radiance file, but for other dates a new file would be required. Details are described in the S5P formaldehyde algorithm documentation.
To optimize the retrieval quality, another advanced configuration option used here is the fitting of the slit function (ISRF) during QDOAS's internal calibration step. An initial estimate of the ISRF is provided in the file data/isrf_band_3_row_225.txt.
B. Python packages
QDOAS is included in the most recent version atmospheric virtual laboratory (avl package), and hence additional installations for QDOAS are not needed. However, you should check that you have the needed qdoas components installed by running conda list in your environment. If your list includes beamf, qdoas and qdoas2harp you are good to go, otherwise avl needs to be updated.
The following Python packages are needed for running this notebook:
harp(version 1.20 or newer)avl(atmosphere-virtual-lab, version 0.5 or newer)eofetch(for downloading of operational TROPOMI HCHO L2 file)
For more info on eofetch see e.g. eofetch README or eofetch example.
import harp
import avl
import eofetch
import os
import urllib
2. Downloading TROPOMI files using eofetch ¶
Needed TROPOMI L1B and L2 files can be downloaded e.g. using eofetch. To be able to perform eofetch.download, you must have environment variables, CDSE_S3_ACCESS and CDSE_S3_SECRET, set as described in in eofetch README.
After setting the credentials, you can download the needed TROPOMI files. Note that the L1B file size is large (>3Gb), and downloading will take some time.
eofetch.download("S5P_OFFL_L1B_RA_BD3_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc")
eofetch.download("S5P_OFFL_L1B_IR_UVN_20230608T005615_20230608T023745_29278_03_020100_20230608T042535.nc")
eofetch.download("S5P_OFFL_L2__HCHO___20230608T193244_20230608T211414_29289_03_020401_20230610T184103.nc")
3. DOAS fit for formaldehyde using QDOAS ¶
To perform the DOAS fit using the doas_cl command, the QDOAS configuration file, TROPOMI L1B input file, and the DOAS-fit output filename need to be defined. The full command is as follows:
!doas_cl -a "HCHO" -c S5P_TROPOMI_HCHO_ISFRfit_qdoasconfig_radasref.xml -f S5P_OFFL_L1B_RA_BD3_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc -o S5P_L2_QDOASSCD_radasref.nc
-a "HCHO"flag specifies which project is used for the QDOAS.- QDOAS configuration file: for this use case we use pre-defined configuration file
S5P_TROPOMI_HCHO_ISFRfit_qdoasconfig_radasref.xmlfor HCHO DOAS fit that is located in this example at the same folder as our notebook. At command line-crefers to the definition of configuration file. The configuration file uses data from thedatafolder, that needs to be downloaded also. - TROPOMI L1B input file:
S5P_OFFL_L1B_RA_BD3_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc, the input is referred with-fat the command line. - QDOAS output file: The QDOAS output file in this use case is named as
S5P_L2_QDOASSCD_radasref.nc, and referred with-oat the command line. - To reduce the computing time, the xml file specified with the
-coption includes specifications to limit the processing to pixels between the[45, 65]degree latitude interval. To undo this, find the<geolocation selected="rectangle">in the xml file and changerectangletonone.
We first download the necessary data files (in case they are not available locally yet):
datafiles = [
'S5P_TROPOMI_HCHO_ISFRfit_qdoasconfig_radasref.xml',
'harp_hcho.json',
'data/O3223_Serdyuchenko(2014)_223K_213-1100nm(2013 version).xs',
'data/O3243_Serdyuchenko(2014)_243K_213-1100nm(2013 version).xs',
'data/bro_Fleischmann(2004)_223K_300-385nm.xs',
'data/ch2o_MellerMoortgat(2000)_298K_224.56-376.00nm(0.01nm).xs',
'data/isrf_band_3_row_225.txt',
'data/no2_VANDAELE_1998_220K.xs',
'data/o3lambda_serdyunchenko_223.xs',
'data/o3squared_serdyunchenko_223.xs',
'data/o4_thalman_volkamer_293K.xs',
'data/rar_20230608.nc',
'data/ring_sao2010_hr_norm.xs',
'data/sao2010_solref_vac.dat',
]
if not os.path.exists('data'):
os.mkdir('data')
data_url = 'https://raw.githubusercontent.com/stcorp/avl-use-cases/master/usecase08/'
for file in datafiles:
if not os.path.exists(file):
urllib.request.urlretrieve(data_url + urllib.parse.quote(file), file)
if os.path.exists('S5P_L2_QDOASSCD_radasref.nc'):
os.remove('S5P_L2_QDOASSCD_radasref.nc')
Then we call qdoas to perform the retrieval.
Note the %%sh at the beginning, which means that the command will be executed as a shell command. The processing of the file can take some time.
%%sh
doas_cl -a "HCHO" -c S5P_TROPOMI_HCHO_ISFRfit_qdoasconfig_radasref.xml -f S5P_OFFL_L1B_RA_BD3_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc -o S5P_L2_QDOASSCD_radasref.nc
Processing file S5P_OFFL_L1B_RA_BD3_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc
Process file S5P_OFFL_L1B_RA_BD3_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc, retCode = 0
4. Converting QDOAS output to HARP compliant format¶
To prepare input for BeAMF, the QDOAS outputfile S5P_L2_QDOASSCD_radasref.nc needs to be converted into a HARP compliant format. For this the qdoas2harp command is used. The command qd2hp performs conversion and it is used as follows:
qd2hp -outdir /path/to/outdir/ -fitwin hcho_analysis -slcol "ch2o=HCHO" S5P_L2_QDOASSCD_radasref.nc
where
-outdiris the path for the QDOAS output file.-fitwinis the fitting window appropriate for HCHO. Here hcho_analysis is used that is given in our QDOAS output file.-slcolrefers to slant column and "ch2o=HCHO" specify which symbol indicates formaldehyde.
Note that in this example all the input and output files are kept in the same folder with this notebook. That is why -outdir . is used in the command below, that refers to the current directory. Now, to run qd2hp in the notebook, you need to add the %%sh at the beginning, before the actual shell command:
%%sh
qd2hp -outdir . -fitwin hcho_analysis -slcol "ch2o=HCHO" S5P_L2_QDOASSCD_radasref.nc
After executing the command succesfully, output file with selected information from the original file in HARP-compliant format is created, that will be the input for BeAMF:
S5P_OFFL_QDOAS_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc
5. Preparing input for BeAMF: adding auxiliary parameters ¶
In AMF calculation we need information e.g. on cloud and surface properties, and hence the next step is to add this auxiliary data to the harp compliant QDOAS file that was created in the previous section. We take this cloud and surface information from the operational S5P L2 HCHO file that corresponds to the example DOAS-fit file we exctracted in the previous step.
You can check which operational S5P L2 HCHO file is needed by using aux_imp command together with the name of the HARP compliant QDOAS file:
%%sh
aux_imp S5P_OFFL_QDOAS_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc
Aux. file that will be used for l2file :
S5P_OFFL_QDOAS_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc ----> S5P_OFFL_L2__HCHO___20230608T193244_20230608T211414_29289_03_*.nc
Now that the L2 file is downloaded already at Sect. 2, the auxiliary parameters can be extracted. After the command is executed succesfully, the auxiliary variables will be written into the HARP compliant QDOAS file (Sect. 3). Note that the extraction of the variables can take a while. As all the files are in the same working folder as the notebook, the extraction can be run as follows (--auxdir . pointing to the working folder):
%%sh
aux_imp S5P_OFFL_QDOAS_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc --auxdir .
The resulting S5P_OFFL_QDOAS file can be read with harp to print out the contents. This data will be part of the input needed to run BeAMF.
qdoas=harp.import_product('S5P_OFFL_QDOAS_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc')
print(qdoas)
source product = 'S5P_OFFL_QDOAS_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc'
history = 'qdoas2harp conversion. {"qdoas file": "S5P_L2_QDOASSCD_radasref.nc", "name of sensor": "TROPOMI", "fitwindow name": "hcho_analysis", "fitwindow range": [328.5, 359.0], "L1 file": "S5P_OFFL_L1B_RA_BD3_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc", "L1 spectral band": "UVIS_3 / BAND3", "QDOAS version": "Qdoas version 3.6.5 - 12 March 2024"}'
float solar_zenith_angle {time=1877850} [degree]
float solar_azimuth_angle {time=1877850} [degree]
float sensor_zenith_angle {time=1877850} [degree]
float sensor_azimuth_angle {time=1877850} [degree]
float latitude {time=1877850} [degree_north]
float longitude {time=1877850} [degree_east]
double datetime_start {time=1877850} [seconds since 2000-01-01]
float longitude_bounds {time=1877850, 4} [degree_east]
float latitude_bounds {time=1877850, 4} [degree_north]
float HCHO_slant_column_number_density {time=1877850} [molecules/cm2]
float HCHO_slant_column_number_density_uncertainty {time=1877850} [molecules/cm2]
long scan_subindex {time=1877850}
float cloud_fraction {time=1877850} []
float cloud_albedo {time=1877850} []
float cloud_albedo_uncertainty {time=1877850} []
float cloud_fraction_uncertainty {time=1877850} []
float cloud_height {time=1877850} [km]
float cloud_height_uncertainty {time=1877850} [km]
float cloud_pressure {time=1877850} [Pa]
float cloud_pressure_uncertainty {time=1877850} [Pa]
float surface_albedo {time=1877850} []
float surface_altitude {time=1877850} [m]
float surface_altitude_uncertainty {time=1877850} [m]
double pressure {time=1877850, vertical=34} [Pa]
float surface_pressure {time=1877850} [Pa]
double tropopause_pressure {time=1877850} [Pa]
float HCHO_volume_mixing_ratio_dry_air_apriori {time=1877850, vertical=34} [ppv]
6. Air Mass Factor calculation with BeAMF ¶
This is the actual step to calculate the AMF. The BeAMF requires several input parameters, that are defined in the configuration file harp_hcho.json. The input parameters include e.g. the Look-Up-Table file (for this case LUT_AMF_I_300nm_500nm_S1_US_small_new.nc) and the QDOAS file that has been prepared in the previous Sections.
The LUT file is retrieved from the cache of example data.
avl.download("LUT_AMF_I_300nm_500nm_S1_US_small_new.nc")
'LUT_AMF_I_300nm_500nm_S1_US_small_new.nc'
The BeAMF tool and AMF calculation is executed as follows (note that this can take several minutes):
%%sh
beamf -c harp_hcho.json
BeAMF tool is running
Running time: 250.69082498550415
Program is end!
The new parameters from BeAMF are again written into the S5P_OFFL_QDOAS - file. By printing out the file we can see the new parameters that have been added, including e.g. HCHO_column_number_density:
hcho=harp.import_product('S5P_OFFL_QDOAS_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc')
print(hcho)
source product = 'S5P_OFFL_QDOAS_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc'
history = 'qdoas2harp conversion. {"qdoas file": "S5P_L2_QDOASSCD_radasref.nc", "name of sensor": "TROPOMI", "fitwindow name": "hcho_analysis", "fitwindow range": [328.5, 359.0], "L1 file": "S5P_OFFL_L1B_RA_BD3_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc", "L1 spectral band": "UVIS_3 / BAND3", "QDOAS version": "Qdoas version 3.6.5 - 12 March 2024"}'
float solar_zenith_angle {time=1877850} [degree]
float solar_azimuth_angle {time=1877850} [degree]
float sensor_zenith_angle {time=1877850} [degree]
float sensor_azimuth_angle {time=1877850} [degree]
float latitude {time=1877850} [degree_north]
float longitude {time=1877850} [degree_east]
double datetime_start {time=1877850} [seconds since 2000-01-01]
float longitude_bounds {time=1877850, 4} [degree_east]
float latitude_bounds {time=1877850, 4} [degree_north]
float HCHO_slant_column_number_density {time=1877850} [molecules/cm2]
float HCHO_slant_column_number_density_uncertainty {time=1877850} [molecules/cm2]
long scan_subindex {time=1877850}
float cloud_fraction {time=1877850} []
float cloud_albedo {time=1877850} []
float cloud_albedo_uncertainty {time=1877850} []
float cloud_fraction_uncertainty {time=1877850} []
float cloud_height {time=1877850} [km]
float cloud_height_uncertainty {time=1877850} [km]
float cloud_pressure {time=1877850} [Pa]
float cloud_pressure_uncertainty {time=1877850} [Pa]
float surface_albedo {time=1877850} []
float surface_altitude {time=1877850} [m]
float surface_altitude_uncertainty {time=1877850} [m]
double pressure {time=1877850, vertical=34} [Pa]
float surface_pressure {time=1877850} [Pa]
double tropopause_pressure {time=1877850} [Pa]
float HCHO_volume_mixing_ratio_dry_air_apriori {time=1877850, vertical=34} [ppv]
float HCHO_column_number_density {time=1877850}
float HCHO_column_number_density_amf {time=1877850}
float tropospheric_HCHO_column_number_density_amf {time=1877850}
float stratospheric_HCHO_column_number_density_amf {time=1877850}
float HCHO_column_number_density_avk {time=1877850, vertical=34}
float cloud_radiance_fraction {time=1877850}
float HCHO_column_number_density_amfgeo {time=1877850}
float HCHO_column_number_density_amfclr {time=1877850}
float HCHO_column_number_density_amfcld {time=1877850}
7. Plotting HCHO data ¶
As the region of interest in this use case is over Canada, for plotting purposes the resulting HCHO data is imported with latitude condition. Over intense fire areas elevated HCHO is clearly visible.
filename='S5P_OFFL_QDOAS_20230608T193244_20230608T211414_29289_03_020100_20230608T230242.nc'
operations="latitude>40;latitude<62"
hcho_product = harp.import_product(filename, operations)
avl.Geo(hcho_product,value="HCHO_column_number_density", colormap="viridis", colorrange=(0.5e15,2e16), centerlat=55, centerlon=-110, zoom=4)